DNAcycP
Center Postdoc Keren Li worked with center Professors Ji-Ping Wang and Alec Wang to create DNAcycP, a new tool to model DNA bendability. For more information, check out this post, or the web platform as well as the stand-alone DNAcycP python code
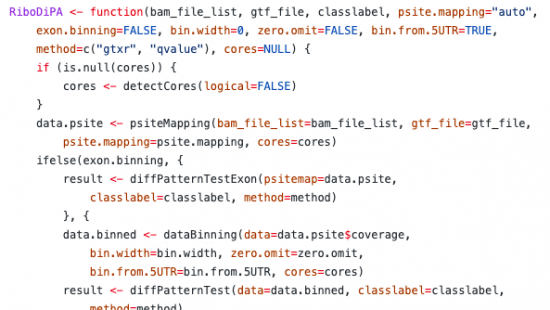
RiboDiPA
RiboDiPA, short for Ribosome Diferential Pattern Analysis, is a bioinformatics pipeline developed for analysis of the pattern of Ribo-seq footprint data. RiboDiPA is released as an R package to support statistical inference of translational differences between conditions. Briefly, this involves mapping Ribo-seq data to P-site counts along a total transcript of a gene, followed by binning these counts and performing bin-wise statistical testing. For more information and code, click here.
TimeCycle
TimeCycle is designed to detect rhythmic genes in circadian transcriptomic time-series data. Based on topological data analysis, TimeCycle provides a reliable and efficient reference-free framework for cycle detection — handling custom sampling schemes, replicates, and missing data. For more information and R source code, click here
TimeTrial
Elan Ness-Cohn and Rosemary Braun worked to create TimeTrial, an open-source software for the design and optimization of circadian transcriptomics experiments. For downloads and more information, click here
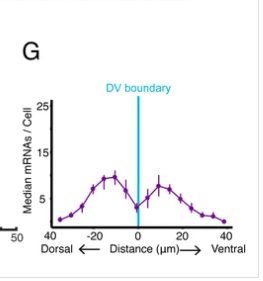
smFISH
Rachael Bakker worked with center investigators Madhav Mani and Richard Carthew to create smFISH, a technique to perform single molecule FISH in Drosophila imaginal discs. For more information, check out their manuscript The Wg and Dpp morphogens regulate gene expression by modulating the frequency of transcriptional bursts. eLife (in press).
Fly-QMA
Center student Sebastian Bernasek worked with Professor Luis Amaral to create Fly-QMA, which helps hto quantify and analyze expression patterns in the Drosophila imaginal disc. For more information, check out their website!